In this post, I explore why NeuroQuant processing uses a 3D T1 sagittal MRI sequence and that using other sequences will generate an unsupported MR sequence error message.
Previously, I reviewed how to ensure accurate NeuroQuant processing results, and my colleague discussed why contrast agents are not used to achieve quality segmentation with NeuroQuant, and examined why good alignment to atlas is necessary for accurate NeuroQuant output.
3D T1 sagittal MRIs are needed to obtain ideal brain morphometry for volume analysis. In order to achieve its high-quality segmentation results, NeuroQuant has been optimized for processing 3D T1 sagittal MRI acquisition sequence in 1.2, 1.5 and 3.0T scanners.
As a quality control feature, NeuroQuant contains a data filtering mechanism that provides verification of study scanning parameters prior to initiating processing. Ensuring proper processing preserves the accuracy of the segmentation results and therefore, the accuracy of the calculated brain volumes. To protect the accuracy of the results, NeuroQuant prevents analysis when sequences other than 3D T1 are sent for processing.
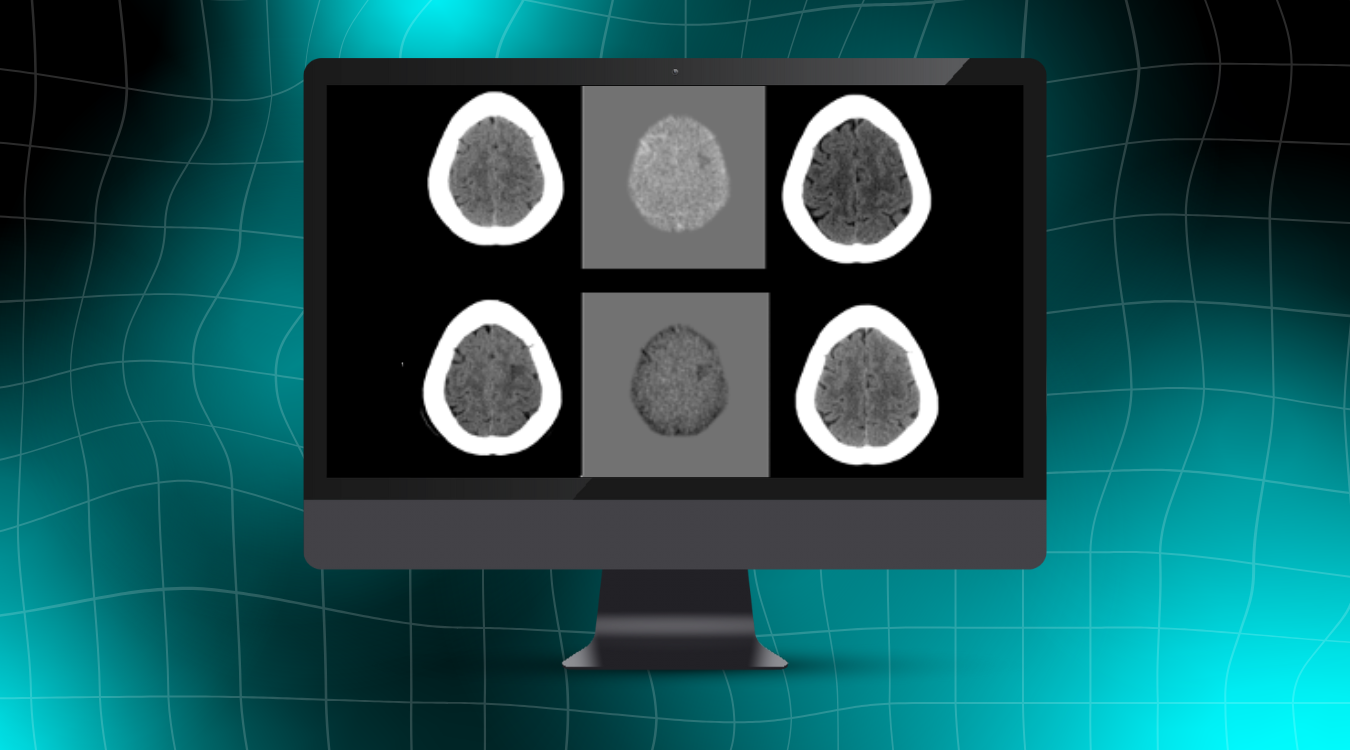
The unsupported sequences provided as examples below will generate an error message.
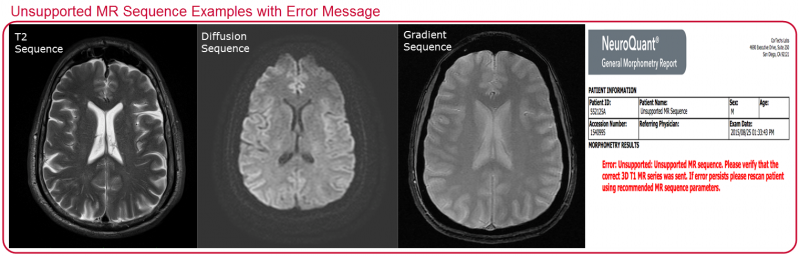
Another example that will generate an error message happens when the wrong series (or entire exam) is sent. When an entire exam is sent instead of just the required 3D T1 NeuroQuant scan, there can be a delay in the return of NeuroQuant results, which will result in multiple error messages being sent back to the PACS.
More information about using and processing NeuroQuant can be found here.